
Cells interact with their surrounding environment by secreting proteins which act as messengers or signals for communicating with other cells. Capturing these elusive and minute proteins, particularly those from our immune cells, and correlating them to the individual source cells can provide important insights into immune responses in patients with chronic and infectious diseases.
For patients with cancer, autoimmune disorders, or infectious diseases, such insights can help accelerate the development of immunotherapies.
However, studying how unhealthy or healthy cells communicate, interact and coordinate with each other in response to stimuli or pathogens, remains a challenge.
To help fill the gap in correlating cell functions with their secreted proteins, a team of researchers led by Assistant Professor Cheow Lih Feng from the Department of Biomedical Engineering at CDE, as well as the NUS Institute for Health Innovation & Technology, has invented a new technique to facilitate the studying of immune cell response at a single-cell level.
The team’s breakthrough, called time-resolved assessment of single-cell protein secretion with sequencing (TRAPS-seq), was reported in Nature Methods earlier this year.
"Having progressive snapshots of the dynamic mechanism of immune cells - from responding to a viral infection, to eliminating the threat - can accelerate the development of new therapeutic strategies for disease treatment, such as targeting mechanisms that are selective for cancer cells and hence spare healthy cells and reduce side effects," said Asst Prof Cheow.
"In addition, a better understanding of how healthy cells communicate can provide valuable insights into the normal cellular processes disrupted in cancer."
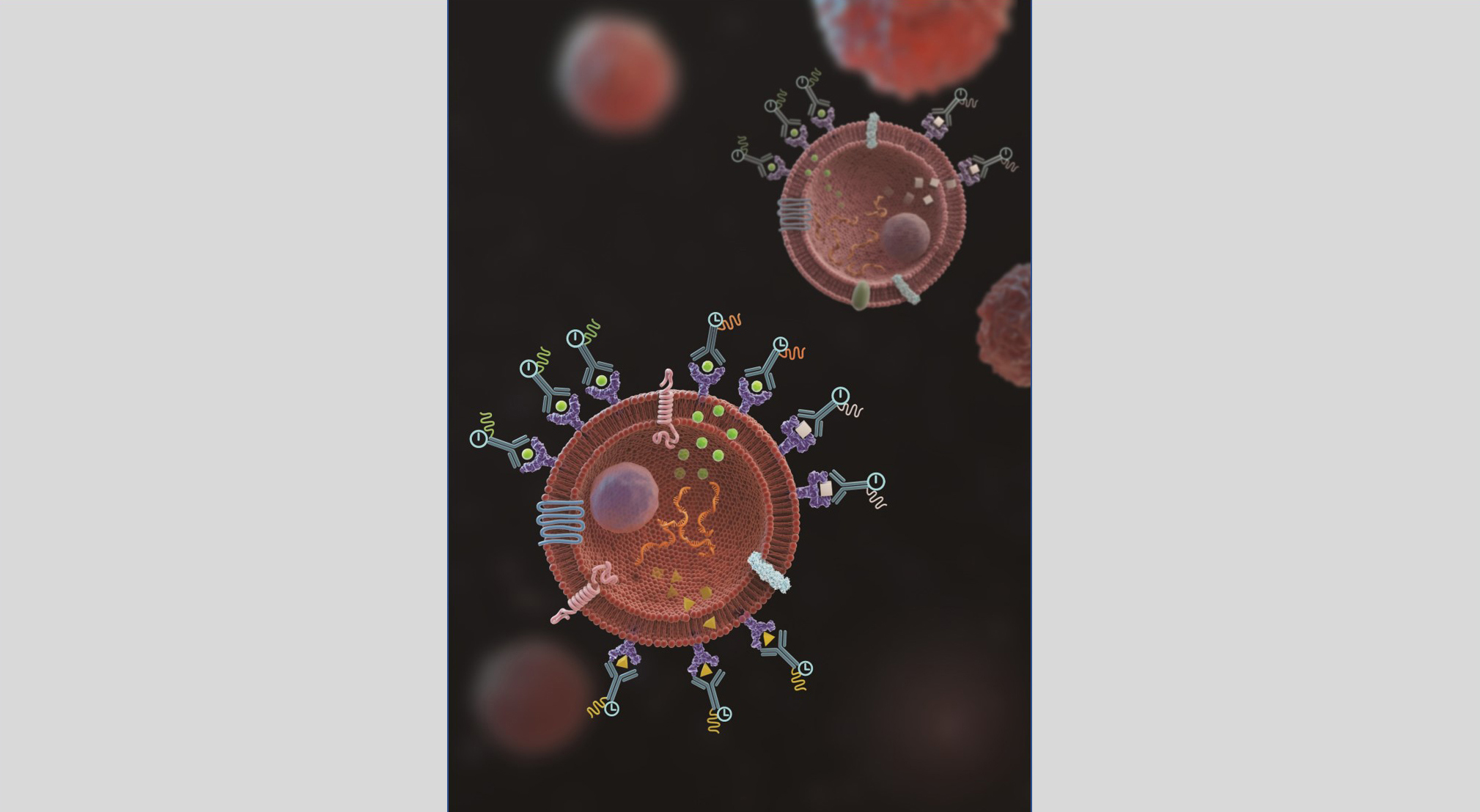
Gathering data on cell functionality
TRAPS-seq employs surface proteins on a cell to anchor the secreted proteins by the cell, which can then be easily analysed by currently available techniques.
The unique advantage of this method over other existing techniques is its capability to track the source cell secreting the proteins and measure the changes in the secretion over time when it responds to a stimulus.
"By modifying the surface proteins on a cell, we can use them as a hook to anchor a secreted protein as soon as it emerges from the cell surface. Using this approach, we can combine with other techniques, such as 10x Genomics barcoding assay, to correlate the cell’s functionality directly with its transcription and surface phenotypic profiles," said Dr Wu Tongjin, lead author of the paper.
To test the technique's viability, the team used TRAPS-seq to simultaneously monitor three types of secreted cytokine proteins from T-cells, a type of white blood cells linked to our immune defence system to fight against various diseases, by exposing them to a stimulus.
The team was able to track the dynamic changes of these secreted proteins over time and observe sub-populations of T-cells that show different dynamic patterns and protein secretion profiles, indicating the potential of using the method to discover new dynamic cellular mechanisms.
"TRAPS-seq provides the opportunity to measure the protein secretion profile of individual cells and resolve their functional differences,” Asst Prof Cheow said.
“Under an abnormal condition, changes to protein secretion would have broad-ranging consequences to our immune system, tissue development and disease progression. The new technique holds great potential as a powerful tool for scientists to discover therapeutic targets for diseases relating to dysregulated protein secretion."
Next steps
Currently, the NUS team can profile three secreted proteins at a time, but the researchers are working towards increasing the number of secreted proteins being studied simultaneously to 10.
The TRAPS-seq assay works for any secreted protein, although immediate applications are likely in immunology profiling. The researchers are optimistic that the extensive catalogues of verified antibody pairs for enzyme-linked immunoassay (ELISA) could be adapted to TRAPS-seq to detect a broad range of secreted proteins for various applications.





